Introduction¶
OpenMiChroM is a Python library for performing chromatin dynamics simulations [1]. OpenMiChroM uses the OpenMM Python API employing the MiChroM (Minimal Chromatin Model) energy function [2]. The chromatin dynamics simulations generate an ensemble of 3D chromosomal structures that are consistent with experimental Hi-C maps [3]. Open-MiChroM also allows simulations of a single or multiple chromosome chain using High-Performance Computing in different platforms (GPUs and CPUs).
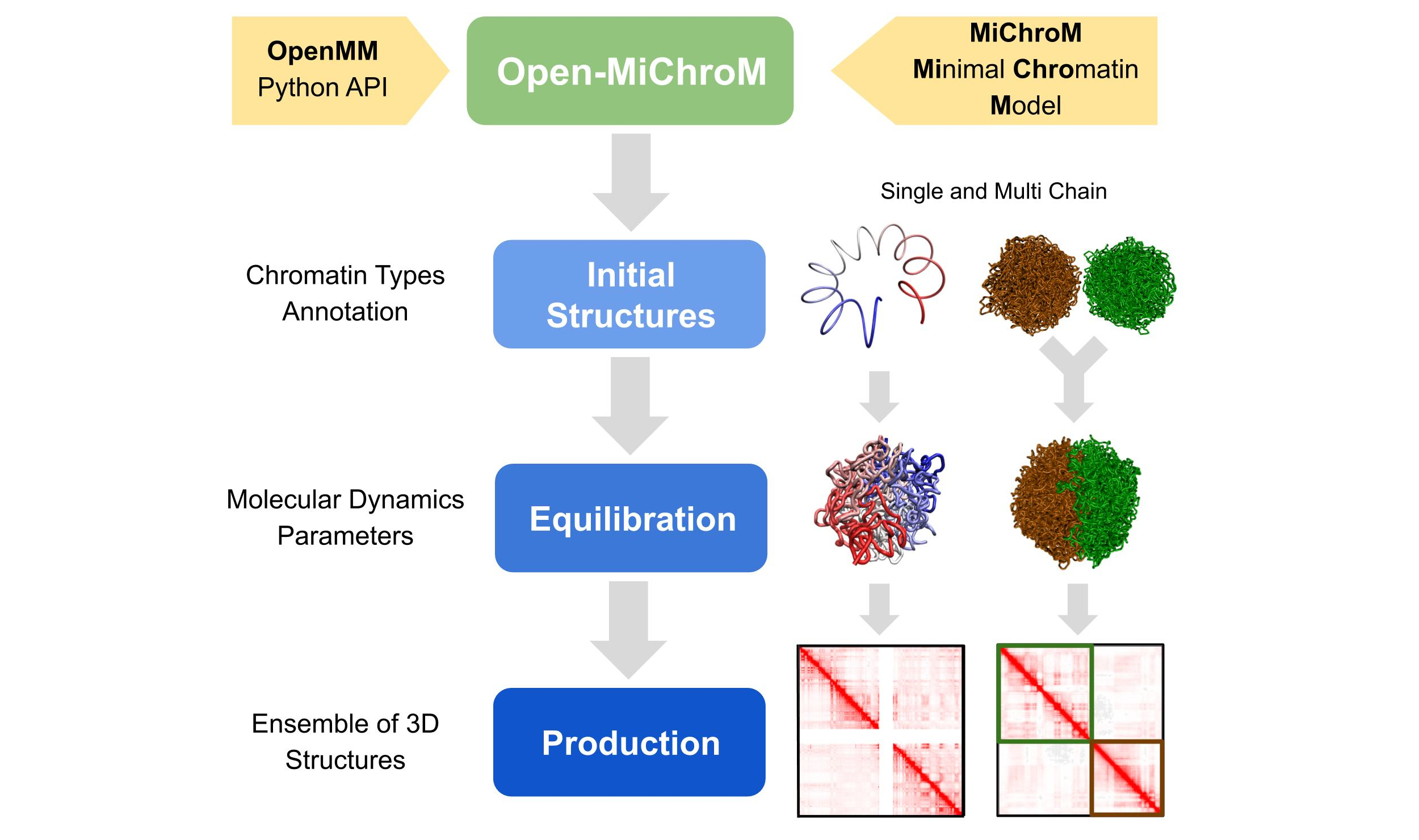
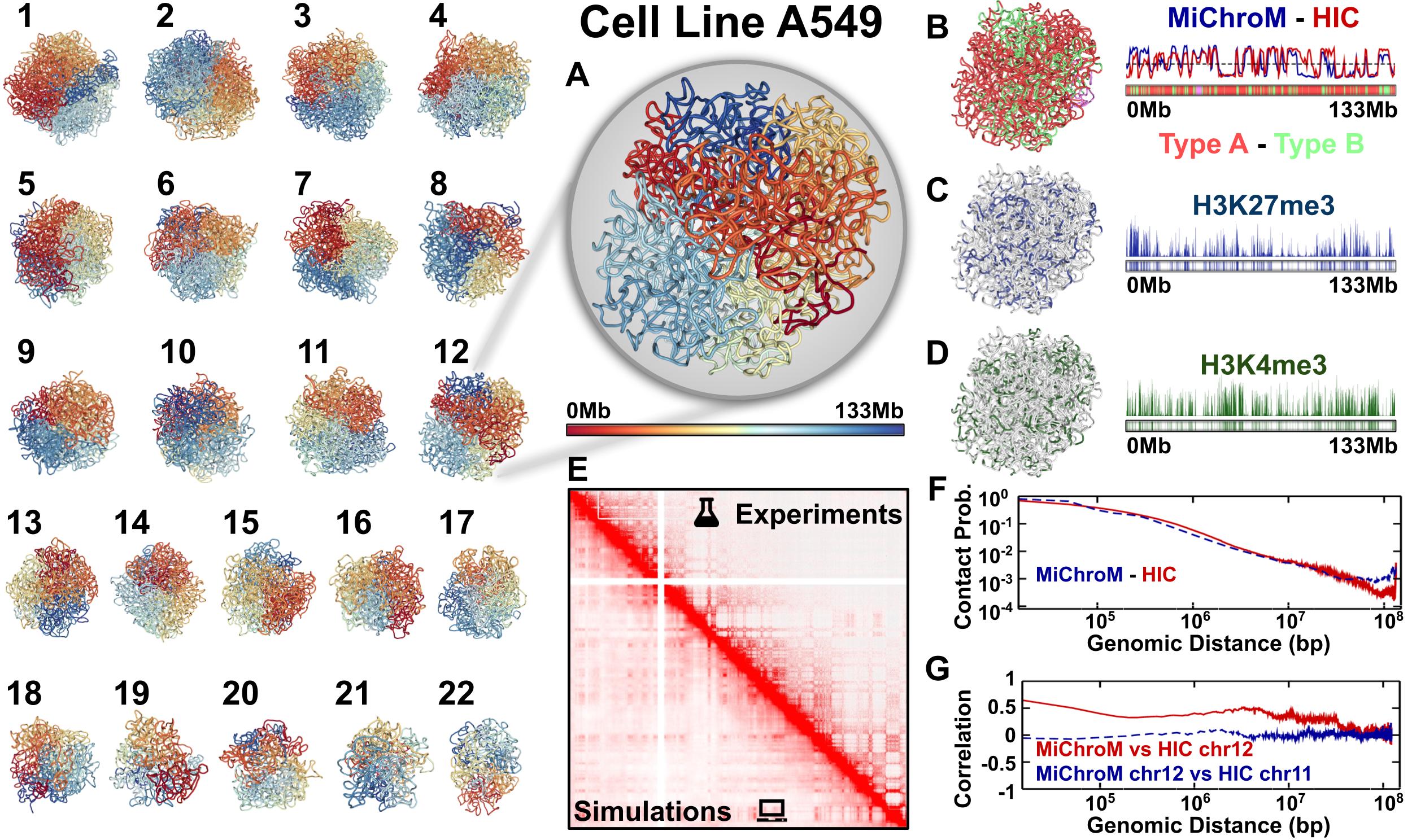
The chromatin dynamics simulations can be performed for different human cell lines, cell phases (interphase to metaphase), and different organisms from DNAzoo. Chromatin subcompartment annotations are available at the NDB (Nucleome Data Bank). OpenMiChroM package receives the chromatin sequence of compartments and subcompartments as input to create and simulate a chromosome polymer model. Examples of running the simulations and generating the in silico Hi-C maps can be found here
- 1
Antonio B. Oliveira Junior, Vinícius G. Contessoto, Matheus F. Mello, and José N. Onuchic. A Scalable Computational Approach for Simulating Complexes of Multiple Chromosomes. Journal of Molecular Biology, 433(6):166700, March 2021. URL: https://linkinghub.elsevier.com/retrieve/pii/S0022283620306185 (visited on 2021-05-15), doi:10.1016/j.jmb.2020.10.034.
- 2
Michele Di Pierro, Bin Zhang, Erez Lieberman Aiden, Peter G. Wolynes, and José N. Onuchic. Transferable model for chromosome architecture. Proceedings of the National Academy of Sciences, 113(43):12168–12173, October 2016. URL: http://www.pnas.org/lookup/doi/10.1073/pnas.1613607113, doi:10.1073/pnas.1613607113.
- 3
Vinícius G Contessoto, Ryan R Cheng, Arya Hajitaheri, Esteban Dodero-Rojas, Matheus F Mello, Erez Lieberman-Aiden, Peter G Wolynes, Michele Di Pierro, and José N Onuchic. The Nucleome Data Bank: web-based resources to simulate and analyze the three-dimensional genome. Nucleic Acids Research, 49(D1):D172–D182, January 2021. URL: https://academic.oup.com/nar/article/49/D1/D172/5918320, doi:10.1093/nar/gkaa818.